cenplot
A Python library for building centromere figures.
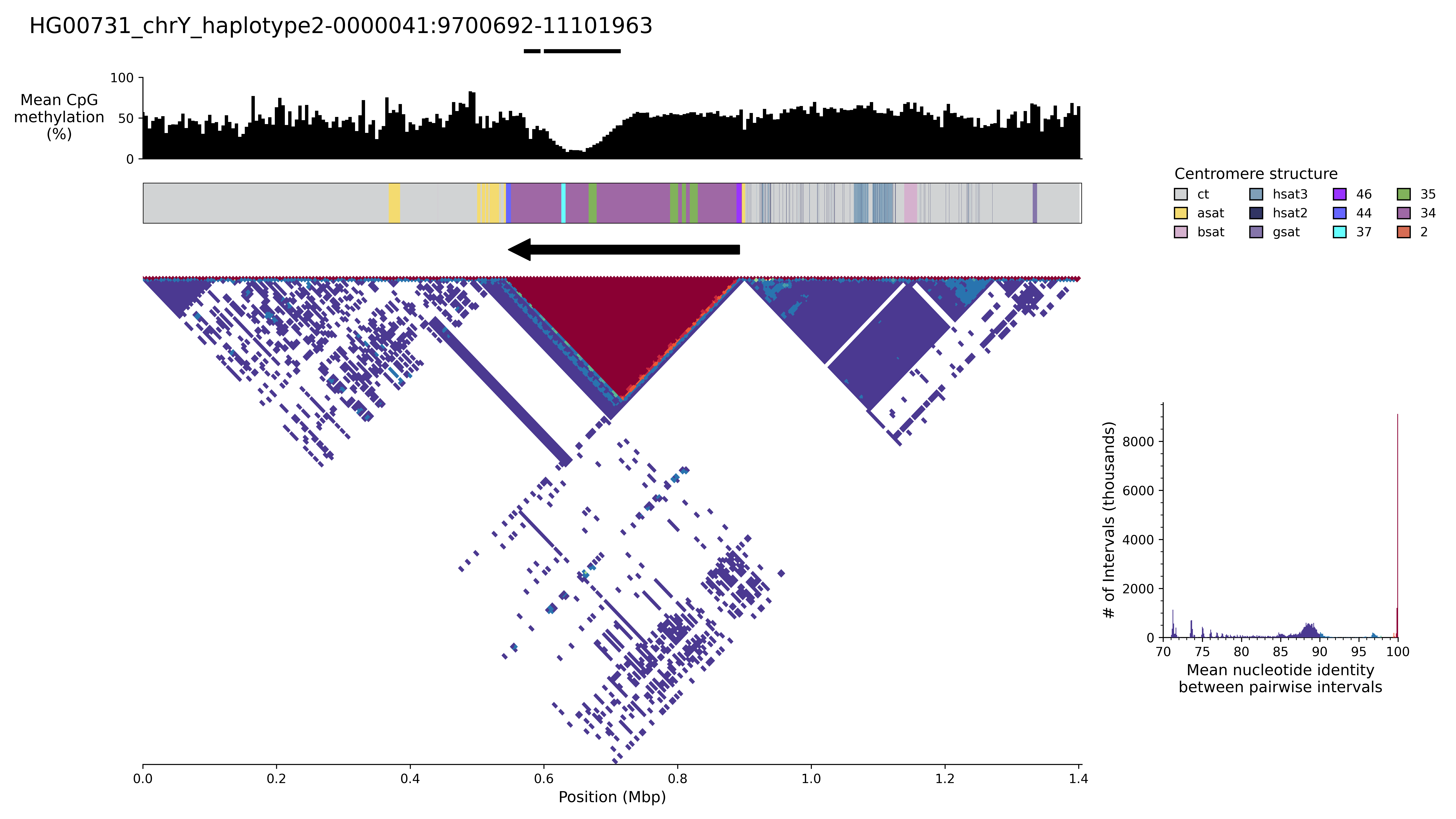
Quickstart
Getting Started
Install the package from pypi.
pip install cenplot
CLI
Generating a split HOR tracks using the cenplot draw command.
# examples/example_cli.sh
cenplot draw \
-t tracks_hor.toml \
-c "chm13_chr10:38568472-42561808" \
-p 4 \
-d plots \
-o "plot/merged_image.png"
Python API
The same HOR track can be created with a few lines of code.
# examples/example_api.py
from cenplot import plot_one_cen, read_one_cen_tracks
chrom = "chm13_chr10:38568472-42561808"
track_list, settings = read_one_cen_tracks("tracks_hor.toml", chrom=chrom)
fig, axes, outfile = plot_one_cen(track_list.tracks, "plots", chrom, settings)
Development
Requires Python >= 3.12 and Git LFS to pull test files.
Create a venv, build cenplot, and install it. Also, generate the docs.
which python3.12
git lfs install && git lfs pull
make dev && make build && make install
pdoc ./cenplot -o docs/
The generated venv will have the cenplot script.
# source venv/bin/activate
venv/bin/cenplot -h
To run tests.
# Takes ~10 minutes
make test
Overview
Configuration comes in the form of TOML files with two fields, [settings] and [[tracks]].
[settings]
format = "png"
[[tracks]]
title = "Alpha-satellite HOR monomers"
position = "relative"
[[tracks]]
title = "Sequence Composition"
position = "relative"
[[settings]] determines figure level settings while [[tracks]] determines track level settings.
- To view all of the possible options for
[[settings]], seecenplot.PlotSettings - To view all of the possible options for
[[tracks]], see one ofcenplot.TrackSettings
Track Order
Order is determined by placement of tracks. Here the "Alpha-satellite HOR monomers" comes before the "Sequence Composition" track.
[[tracks]]
title = "Alpha-satellite HOR monomers"
position = "relative"
[[tracks]]
title = "Sequence Composition"
position = "relative"
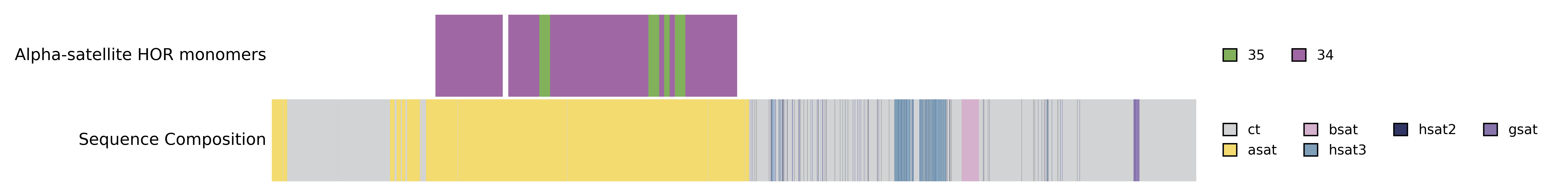
Reversing this will plot "Sequence Composition" before "Alpha-satellite HOR monomers"
[[tracks]]
title = "Sequence Composition"
position = "relative"
[[tracks]]
title = "Alpha-satellite HOR monomers"
position = "relative"

Overlap
Tracks can be overlapped with the position or cenplot.TrackPosition setting.
[[tracks]]
title = "Sequence Composition"
position = "relative"
[[tracks]]
title = "Alpha-satellite HOR monomers"
position = "overlap"
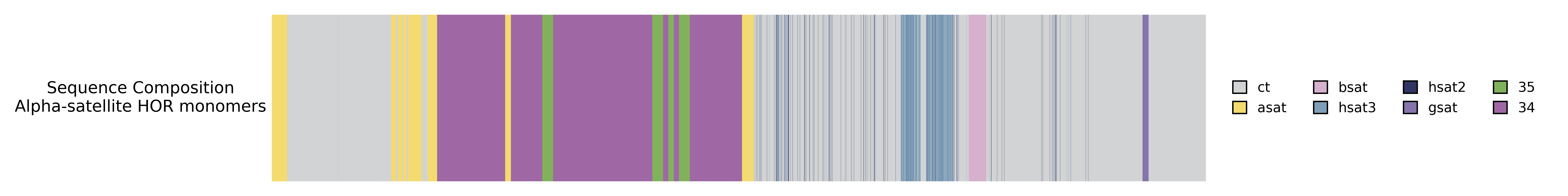
The preceding track is overlapped and the legend elements are merged.
Track Types and Data
Track types, or cenplot.TrackTypes, are specified via the type parameter.
[[tracks]]
title = "Sequence Composition"
position = "relative"
type = "label"
path = "rm.bed"
Each type will expect different BED files in the path option.
- For example, the option
TrackType.SelfIdentexpects the following values.
| query | query_st | query_end | reference | reference_st | reference_end | percent_identity_by_events |
|---|---|---|---|---|---|---|
| x | 1 | 5000 | x | 1 | 5000 | 100.0 |
When using the Python API, each will have an associated read_* function (ex. cenplot.read_bed_identity).
- Using
cenplot.read_one_cen_tracksis preferred.
If input BED files have contigs with coordinates in their name, the coordinates are expected to be in absolute coordinates.
Absolute coordinates
| chrom | chrom_st | chrom_end |
|---|---|---|
| chm13:100-200 | 105 | 130 |
Proportion
Each track must account for some proportion of the total plot dimensions.
- The plot dimensions are specified with
cenplot.PlotSettings.dim
Here, with a total proportion of 0.2, each track will take up 50% of the total plot dimensions.
[[tracks]]
title = "Sequence Composition"
position = "relative"
type = "label"
proportion = 0.1
path = "rm.bed"
[[tracks]]
title = "Alpha-satellite HOR monomers"
position = "relative"
type = "hor"
proportion = 0.1
path = "stv_row.bed"
When the position is cenplot.TrackPosition.Overlap, the proportion is ignored.
[[tracks]]
title = "Sequence Composition"
position = "relative"
type = "label"
proportion = 0.1
path = "rm.bed"
[[tracks]]
title = "Alpha-satellite HOR monomers"
position = "overlap"
type = "hor"
path = "stv_row.bed"
Options
Options for specific cenplot.TrackType types can be specified in options.
[[tracks]]
title = "Sequence Composition"
position = "relative"
proportion = 0.5
type = "label"
path = "rm.bed"
# Both need to be false to keep x
options = { hide_x = false }
[[tracks]]
title = "Alpha-satellite HOR monomers"
position = "overlap"
type = "hor"
path = "stv_row.bed"
# Change mode to showing HOR variant and reduce legend number of cols.
options = { hide_x = false, mode = "hor", legend_ncols = 2 }
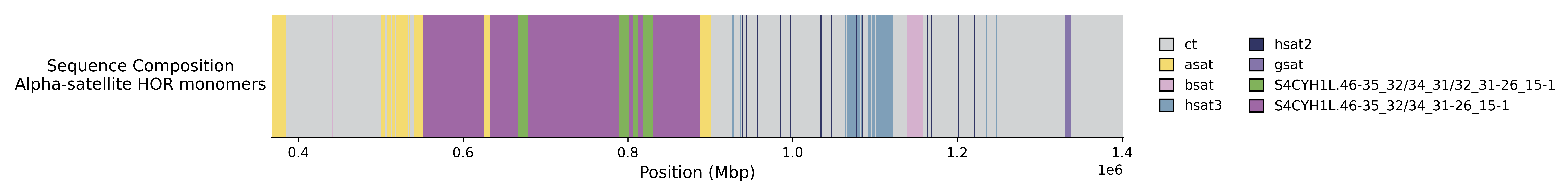
Subset
To subset to a given region, provide the chromosome name with start and end coordinates.
cenplot draw -t track.toml -c "chrom:st-end" -d .
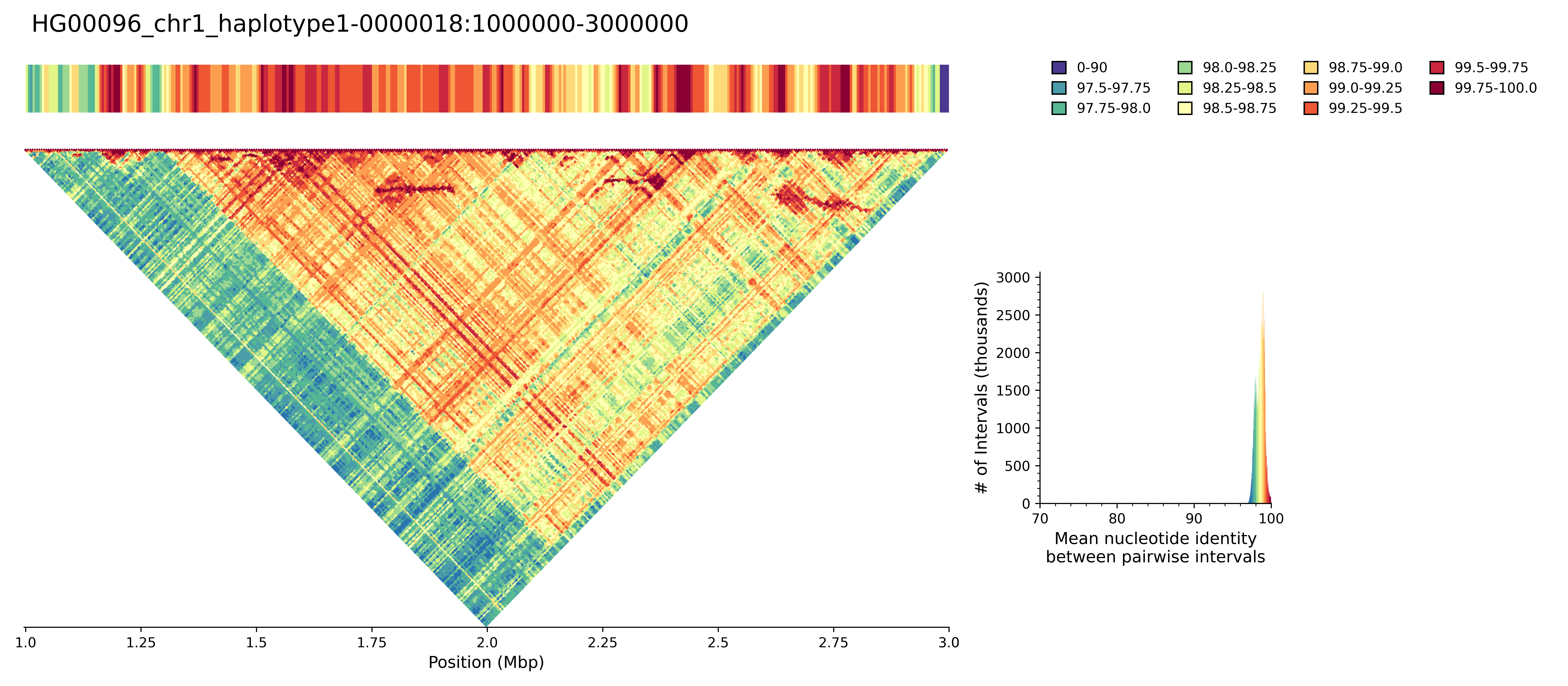
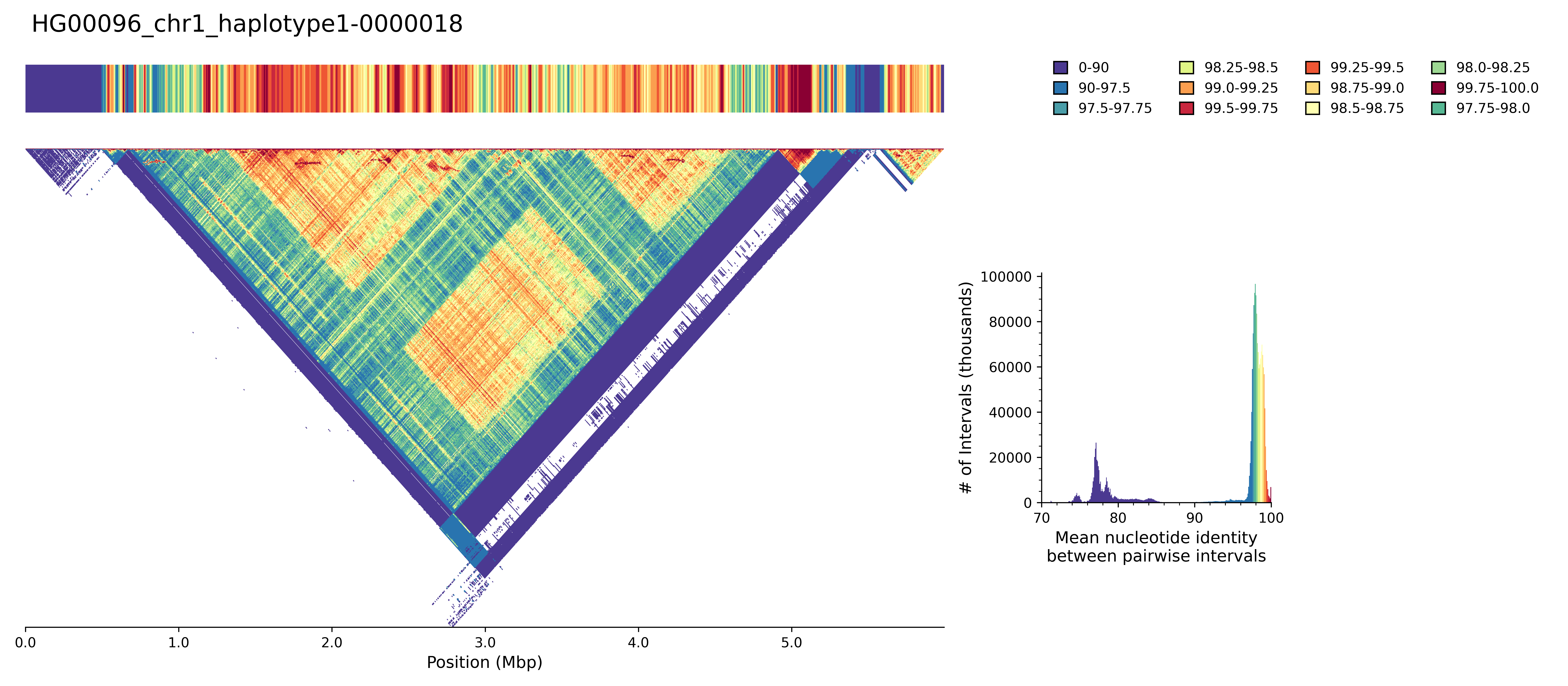
|
Coordinates already existing in the chrom name will be ignored
Examples
Examples of both the CLI and Python API can be found in the root of cenplot's project directory under examples/ or test/
1r""" 2[](https://pypi.org/project/cenplot/) 3[](https://github.com/logsdon-lab/cenplot/actions/workflows/main.yaml) 4[](https://github.com/logsdon-lab/cenplot/actions/workflows/docs.yaml) 5 6A Python library for building centromere figures. 7 8<figure float="left"> 9 <img align="middle" src="https://raw.githubusercontent.com/logsdon-lab/cenplot/refs/heads/main/docs/example_multiple.png" width="100%"> 10</figure> 11 12# Quickstart 13 14.. include:: ../docs/quickstart.md 15 16 17# Overview 18Configuration comes in the form of `TOML` files with two fields, `[settings]` and `[[tracks]]`. 19```toml 20[settings] 21format = "png" 22 23[[tracks]] 24title = "Alpha-satellite HOR monomers" 25position = "relative" 26 27[[tracks]] 28title = "Sequence Composition" 29position = "relative" 30``` 31 32`[[settings]]` determines figure level settings while `[[tracks]]` determines track level settings. 33* To view all of the possible options for `[[settings]]`, see `cenplot.PlotSettings` 34* To view all of the possible options for `[[tracks]]`, see one of `cenplot.TrackSettings` 35 36## Track Order 37Order is determined by placement of tracks. Here the `"Alpha-satellite HOR monomers"` comes before the `"Sequence Composition"` track. 38```toml 39[[tracks]] 40title = "Alpha-satellite HOR monomers" 41position = "relative" 42 43[[tracks]] 44title = "Sequence Composition" 45position = "relative" 46``` 47 48<figure float="left"> 49 <img align="middle" src="https://raw.githubusercontent.com/logsdon-lab/cenplot/refs/heads/main/docs/simple_hor_top.png" width="100%"> 50</figure> 51 52Reversing this will plot `"Sequence Composition"` before `"Alpha-satellite HOR monomers"` 53 54```toml 55[[tracks]] 56title = "Sequence Composition" 57position = "relative" 58 59[[tracks]] 60title = "Alpha-satellite HOR monomers" 61position = "relative" 62``` 63 64<figure float="left"> 65 <img align="middle" src="https://raw.githubusercontent.com/logsdon-lab/cenplot/refs/heads/main/docs/simple_hor_bottom.png" width="100%"> 66</figure> 67 68## Overlap 69Tracks can be overlapped with the `position` or `cenplot.TrackPosition` setting. 70 71```toml 72[[tracks]] 73title = "Sequence Composition" 74position = "relative" 75 76[[tracks]] 77title = "Alpha-satellite HOR monomers" 78position = "overlap" 79``` 80 81<figure float="left"> 82 <img align="middle" src="https://raw.githubusercontent.com/logsdon-lab/cenplot/refs/heads/main/docs/simple_hor_overlap.png" width="100%"> 83</figure> 84 85The preceding track is overlapped and the legend elements are merged. 86 87## Track Types and Data 88Track types, or `cenplot.TrackType`s, are specified via the `type` parameter. 89```toml 90[[tracks]] 91title = "Sequence Composition" 92position = "relative" 93type = "label" 94path = "rm.bed" 95``` 96 97Each type will expect different BED files in the `path` option. 98* For example, the option `TrackType.SelfIdent` expects the following values. 99 100|query|query_st|query_end|reference|reference_st|reference_end|percent_identity_by_events| 101|-|-|-|-|-|-|-| 102|x|1|5000|x|1|5000|100.0| 103 104When using the `Python` API, each will have an associated `read_*` function (ex. `cenplot.read_bed_identity`). 105* Using `cenplot.read_one_cen_tracks` is preferred. 106 107> [!NOTE] If input BED files have contigs with coordinates in their name, the coordinates are expected to be in absolute coordinates. 108 109Absolute coordinates 110|chrom|chrom_st|chrom_end| 111|-|-|-| 112|chm13:100-200|105|130| 113 114## Proportion 115Each track must account for some proportion of the total plot dimensions. 116* The plot dimensions are specified with `cenplot.PlotSettings.dim` 117 118Here, with a total proportion of `0.2`, each track will take up `50%` of the total plot dimensions. 119```toml 120[[tracks]] 121title = "Sequence Composition" 122position = "relative" 123type = "label" 124proportion = 0.1 125path = "rm.bed" 126 127[[tracks]] 128title = "Alpha-satellite HOR monomers" 129position = "relative" 130type = "hor" 131proportion = 0.1 132path = "stv_row.bed" 133``` 134 135When the position is `cenplot.TrackPosition.Overlap`, the proportion is ignored. 136```toml 137[[tracks]] 138title = "Sequence Composition" 139position = "relative" 140type = "label" 141proportion = 0.1 142path = "rm.bed" 143 144[[tracks]] 145title = "Alpha-satellite HOR monomers" 146position = "overlap" 147type = "hor" 148path = "stv_row.bed" 149``` 150 151## Options 152Options for specific `cenplot.TrackType` types can be specified in `options`. 153* See `cenplot.TrackSettings` 154 155```toml 156[[tracks]] 157title = "Sequence Composition" 158position = "relative" 159proportion = 0.5 160type = "label" 161path = "rm.bed" 162# Both need to be false to keep x 163options = { hide_x = false } 164 165[[tracks]] 166title = "Alpha-satellite HOR monomers" 167position = "overlap" 168type = "hor" 169path = "stv_row.bed" 170# Change mode to showing HOR variant and reduce legend number of cols. 171options = { hide_x = false, mode = "hor", legend_ncols = 2 } 172``` 173 174<figure float="left"> 175 <img align="middle" src="https://raw.githubusercontent.com/logsdon-lab/cenplot/refs/heads/main/docs/simple_hor_track_options.png" width="100%"> 176</figure> 177 178## Subset 179To subset to a given region, provide the chromosome name with start and end coordinates. 180```bash 181cenplot draw -t track.toml -c "chrom:st-end" -d . 182``` 183<table> 184 <tr> 185 <td> 186 <figure float="left"> 187 <img align="middle" src="https://raw.githubusercontent.com/logsdon-lab/cenplot/refs/heads/main/docs/examples_subset.png" width="100%"> 188 </figure> 189 <figure float="left"> 190 <img align="middle" src="https://raw.githubusercontent.com/logsdon-lab/cenplot/refs/heads/main/docs/examples_no_subset.png" width="100%"> 191 </figure> 192 </td> 193 </tr> 194</table> 195 196> [!NOTE] Coordinates already existing in the chrom name will be ignored 197 198## Examples 199Examples of both the CLI and Python API can be found in the root of `cenplot`'s project directory under `examples/` or `test/` 200 201--- 202""" 203 204import logging 205 206from .lib.draw import ( 207 draw_hor, 208 draw_hor_ort, 209 draw_label, 210 draw_strand, 211 draw_self_ident, 212 draw_bar, 213 draw_line, 214 draw_legend, 215 draw_self_ident_hist, 216 draw_local_self_ident, 217 plot_tracks, 218 merge_plots, 219 PlotSettings, 220) 221from .lib.io import ( 222 read_bed9, 223 read_bed_hor, 224 read_bed_identity, 225 read_bed_label, 226 read_track, 227 read_tracks, 228) 229from .lib.track import ( 230 Track, 231 TrackType, 232 TrackPosition, 233 TrackList, 234 LegendPosition, 235 TrackSettings, 236 SelfIdentTrackSettings, 237 LineTrackSettings, 238 LocalSelfIdentTrackSettings, 239 HORTrackSettings, 240 HOROrtTrackSettings, 241 StrandTrackSettings, 242 BarTrackSettings, 243 LabelTrackSettings, 244 PositionTrackSettings, 245 LegendTrackSettings, 246 SpacerTrackSettings, 247) 248 249__author__ = "Keith Oshima (oshimak@pennmedicine.upenn.edu)" 250__license__ = "MIT" 251__all__ = [ 252 "plot_tracks", 253 "merge_plots", 254 "draw_hor", 255 "draw_hor_ort", 256 "draw_label", 257 "draw_self_ident", 258 "draw_self_ident_hist", 259 "draw_local_self_ident", 260 "draw_bar", 261 "draw_line", 262 "draw_strand", 263 "draw_legend", 264 "read_bed9", 265 "read_bed_hor", 266 "read_bed_identity", 267 "read_bed_label", 268 "read_track", 269 "read_tracks", 270 "Track", 271 "TrackType", 272 "TrackPosition", 273 "TrackList", 274 "LegendPosition", 275 "PlotSettings", 276 "TrackSettings", 277 "SelfIdentTrackSettings", 278 "LocalSelfIdentTrackSettings", 279 "StrandTrackSettings", 280 "HORTrackSettings", 281 "HOROrtTrackSettings", 282 "BarTrackSettings", 283 "LineTrackSettings", 284 "LabelTrackSettings", 285 "PositionTrackSettings", 286 "LegendTrackSettings", 287 "SpacerTrackSettings", 288] 289 290logging.getLogger(__name__).addHandler(logging.NullHandler())
24def plot_tracks( 25 tracks: list[Track], 26 settings: PlotSettings, 27 outdir: str | None = None, 28 chrom: str | None = None, 29) -> tuple[Figure, np.ndarray, list[str]]: 30 """ 31 Plot a single centromere figure from a list of `Track`s. 32 33 # Args 34 * `tracks` 35 * List of tracks to plot. The order in the list determines placement on the figure. 36 * `settings` 37 * Settings for output plots. 38 * `outdir` 39 * Output directory. 40 * If not provided, does not output files. 41 * `chrom` 42 * Chromosome label. Replaces {chrom} format string in title if provided and sets output filenames. 43 * If not provided, defaults to "out". 44 45 # Returns 46 * Figure, its axes, and the output filename(s). 47 48 # Usage 49 ```python 50 import cenplot 51 52 chrom = "chm13_chr10:38568472-42561808" 53 track_list, settings = cenplot.read_tracks("tracks_example_api.toml", chrom=chrom) 54 fig, axes, _ = cenplot.plot_tracks(track_list.tracks, settings) 55 ``` 56 """ 57 # Show chrom trimmed of spaces for logs and filenames. 58 logging.info(f"Plotting {len(tracks)} tracks.") 59 60 if not settings.xlim: 61 # Get min and max position of all tracks for this cen. 62 _, min_st_pos = get_min_max_track(tracks, typ="min") 63 _, max_end_pos = get_min_max_track(tracks, typ="max", default_col="chrom_end") 64 else: 65 min_st_pos = settings.xlim[0] 66 max_end_pos = settings.xlim[1] 67 68 # # Scale height based on track length. 69 # adj_height = height * (trk_max_end / max_end_pos) 70 # height = height if adj_height == 0 else adj_height 71 72 fig, axes, track_indices = create_subplots( 73 tracks, 74 settings, 75 ) 76 if settings.legend_pos == LegendPosition.Left: 77 track_col, legend_col = 1, 0 78 else: 79 track_col, legend_col = 0, 1 80 81 track_labels: list[str] = [] 82 83 def get_track_label( 84 chrom: str | None, track: Track, all_track_labels: list[str] 85 ) -> str: 86 if not track.title or not chrom: 87 return "" 88 try: 89 fmt_track_label = track.title.format(chrom=chrom) 90 except KeyError: 91 fmt_track_label = track.title 92 93 track_label = fmt_track_label.encode("ascii", "ignore").decode("unicode_escape") 94 95 # Update track label for each overlap. 96 if track.pos == TrackPosition.Overlap: 97 try: 98 track_label = f"{all_track_labels[-1]}\n{track_label}" 99 except IndexError: 100 pass 101 102 return track_label 103 104 num_hor_split = 0 105 legend_tracks: list[tuple[Axes, Track]] = [] 106 for idx, track in enumerate(tracks): 107 track_row = track_indices[idx] 108 track_label = get_track_label(chrom, track, track_labels) 109 # Store label if more overlaps. 110 track_labels.append(track_label) 111 112 try: 113 track_ax: Axes = axes[track_row, track_col] 114 except IndexError: 115 print(f"Cannot get track ({track_row, track_col}) for {track}.") 116 continue 117 try: 118 legend_ax: Axes | None = axes[track_row, legend_col] 119 except IndexError: 120 legend_ax = None 121 122 # Set xaxis limits 123 track_ax.set_xlim(min_st_pos, max_end_pos) 124 125 # Set labels for both x and y axis. 126 set_both_labels(track_label, track_ax, track) 127 128 if legend_ax: 129 # Make legend title invisible for HORs split after 1. 130 if track.opt == TrackType.HORSplit: 131 legend_ax_legend = legend_ax.get_legend() 132 if legend_ax_legend and num_hor_split != 0: 133 legend_title = legend_ax_legend.get_title() 134 legend_title.set_alpha(0.0) 135 num_hor_split += 1 136 137 # Minimalize all legend cols except self-ident 138 if track.opt != TrackType.SelfIdent or ( 139 track.opt == TrackType.SelfIdent and not track.options.legend 140 ): 141 format_ax( 142 legend_ax, 143 grid=True, 144 xticks=True, 145 xticklabel_fontsize=track.options.legend_fontsize, 146 yticks=True, 147 yticklabel_fontsize=track.options.legend_fontsize, 148 spines=("right", "left", "top", "bottom"), 149 ) 150 else: 151 format_ax( 152 legend_ax, 153 grid=True, 154 xticklabel_fontsize=track.options.legend_fontsize, 155 yticklabel_fontsize=track.options.legend_fontsize, 156 spines=("right", "top"), 157 ) 158 159 if track.opt == TrackType.Legend: 160 # Draw after everything else. 161 legend_tracks.append((track_ax, track)) 162 elif track.opt == TrackType.Position: 163 # Hide everything but x-axis 164 format_ax( 165 track_ax, 166 grid=True, 167 xticklabel_fontsize=track.options.legend_fontsize, 168 yticks=True, 169 yticklabel_fontsize=track.options.legend_fontsize, 170 spines=("right", "left", "top"), 171 ) 172 elif track.opt == TrackType.Spacer: 173 # Hide everything. 174 format_ax( 175 track_ax, 176 grid=True, 177 xticks=True, 178 yticks=True, 179 spines=("right", "left", "top", "bottom"), 180 ) 181 else: 182 # Switch track option. {bar, label, ident, hor} 183 # Add legend. 184 if track.opt == TrackType.HOR or track.opt == TrackType.HORSplit: 185 draw_fn = draw_hor 186 elif track.opt == TrackType.HOROrt: 187 draw_fn = draw_hor_ort 188 elif track.opt == TrackType.Label: 189 draw_fn = draw_label 190 elif track.opt == TrackType.SelfIdent: 191 draw_fn = draw_self_ident 192 elif track.opt == TrackType.LocalSelfIdent: 193 draw_fn = draw_local_self_ident 194 elif track.opt == TrackType.Bar: 195 draw_fn = draw_bar 196 elif track.opt == TrackType.Line: 197 draw_fn = draw_line 198 elif track.opt == TrackType.Strand: 199 draw_fn = draw_strand 200 else: 201 raise ValueError("Invalid TrackType. Unreachable.") 202 203 draw_fn( 204 ax=track_ax, 205 legend_ax=legend_ax, 206 track=track, 207 zorder=idx, 208 ) 209 210 # Draw after all elements added. 211 for ax, track_legend in legend_tracks: 212 draw_legend(ax, axes, track_legend, track_col) 213 214 # Add title 215 if settings.title: 216 if chrom: 217 title = settings.title.format(chrom=chrom) 218 else: 219 title = settings.title 220 fig.suptitle( 221 title, 222 x=settings.title_x, 223 y=settings.title_y, 224 horizontalalignment=settings.title_horizontalalignment, 225 fontsize=settings.title_fontsize, 226 ) 227 # Pad between axes. 228 fig.set_layout_engine(layout=settings.layout, h_pad=settings.axis_h_pad) 229 230 outfiles = [] 231 232 if outdir: 233 os.makedirs(outdir, exist_ok=True) 234 if isinstance(settings.format, str): 235 output_format = [settings.format] 236 else: 237 output_format = settings.format 238 239 # PNG must always be plotted last. 240 # Matplotlib modifies figure settings causing formatting errors in vectorized image formats (svg, pdf) 241 png_output = "png" in output_format 242 if png_output: 243 output_format.remove("png") 244 245 fname = chrom if chrom else "out" 246 for fmt in output_format: 247 outfile = os.path.join(outdir, f"{fname}.{fmt}") 248 fig.savefig(outfile, dpi=settings.dpi, transparent=settings.transparent) 249 outfiles.append(outfile) 250 251 if png_output: 252 outfile = os.path.join(outdir, f"{fname}.png") 253 fig.savefig( 254 outfile, 255 dpi=settings.dpi, 256 transparent=settings.transparent, 257 ) 258 outfiles.append(outfile) 259 260 return fig, axes, outfiles
Plot a single centromere figure from a list of Tracks.
Args
tracks- List of tracks to plot. The order in the list determines placement on the figure.
settings- Settings for output plots.
outdir- Output directory.
- If not provided, does not output files.
chrom- Chromosome label. Replaces {chrom} format string in title if provided and sets output filenames.
- If not provided, defaults to "out".
Returns
- Figure, its axes, and the output filename(s).
Usage
import cenplot
chrom = "chm13_chr10:38568472-42561808"
track_list, settings = cenplot.read_tracks("tracks_example_api.toml", chrom=chrom)
fig, axes, _ = cenplot.plot_tracks(track_list.tracks, settings)
111def merge_plots( 112 figures: list[tuple[Figure, np.ndarray, list[str]]], outfile: str 113) -> None: 114 """ 115 Merge plots produced by `plot_one_cen`. 116 117 # Args 118 * `figures` 119 * List of figures, their axes, and the name of the output files. Only pngs are concatentated. 120 * `outfile` 121 * Output merged file. 122 * Either `png` or `pdf` 123 124 # Returns 125 * None 126 """ 127 if outfile.endswith(".pdf"): 128 with PdfPages(outfile) as pdf: 129 for fig, _, _ in figures: 130 pdf.savefig(fig) 131 else: 132 merged_images = np.concatenate( 133 [ 134 plt.imread(file) 135 for _, _, files in figures 136 for file in files 137 if file.endswith("png") 138 ] 139 ) 140 plt.imsave(outfile, merged_images)
Merge plots produced by plot_one_cen.
Args
figures- List of figures, their axes, and the name of the output files. Only pngs are concatentated.
outfile- Output merged file.
- Either
pngorpdf
Returns
- None
24def draw_hor( 25 ax: Axes, 26 track: Track, 27 *, 28 zorder: float = 1.0, 29 legend_ax: Axes | None = None, 30): 31 """ 32 Draw HOR plot on axis with the given `Track`. 33 """ 34 hide_x = track.options.hide_x 35 legend = track.options.legend 36 border = track.options.bg_border 37 bg_color = track.options.bg_color 38 39 if track.pos != TrackPosition.Overlap: 40 spines = ( 41 ("right", "left", "top", "bottom") if hide_x else ("right", "left", "top") 42 ) 43 else: 44 spines = None 45 46 format_ax( 47 ax, 48 xticks=hide_x, 49 xticklabel_fontsize=track.options.fontsize, 50 yticks=True, 51 yticklabel_fontsize=track.options.fontsize, 52 spines=spines, 53 ) 54 55 ylim = ax.get_ylim() 56 height = ylim[1] - ylim[0] 57 58 if track.options.mode == "hor": 59 colname = "name" 60 else: 61 colname = "mer" 62 63 # Add HOR track. 64 for row in track.data.iter_rows(named=True): 65 start = row["chrom_st"] 66 end = row["chrom_end"] 67 color = row["color"] 68 rect = Rectangle( 69 (start, 0), 70 end + 1 - start, 71 height, 72 color=color, 73 lw=0, 74 label=row[colname], 75 zorder=zorder, 76 ) 77 ax.add_patch(rect) 78 79 if border: 80 # Ensure border is always on top. 81 add_rect(ax, height, zorder + 1.0) 82 83 if bg_color: 84 # Ensure bg is below everything. 85 add_rect(ax, height, zorder - 1.0, fill=True, color=bg_color) 86 87 if legend_ax and legend: 88 draw_uniq_entry_legend( 89 legend_ax, 90 track, 91 ref_ax=ax, 92 ncols=track.options.legend_ncols, 93 loc="center left", 94 alignment="left", 95 )
Draw HOR plot on axis with the given Track.
11def draw_hor_ort( 12 ax: Axes, 13 track: Track, 14 *, 15 zorder: float = 1.0, 16 legend_ax: Axes | None = None, 17): 18 """ 19 Draw HOR ort plot on axis with the given `Track`. 20 """ 21 draw_strand(ax, track, zorder=zorder, legend_ax=legend_ax)
Draw HOR ort plot on axis with the given Track.
10def draw_label( 11 ax: Axes, 12 track: Track, 13 *, 14 zorder: float = 1.0, 15 legend_ax: Axes | None = None, 16) -> None: 17 """ 18 Draw label plot on axis with the given `Track`. 19 """ 20 hide_x = track.options.hide_x 21 color = track.options.color 22 alpha = track.options.alpha 23 legend = track.options.legend 24 border = track.options.bg_border 25 edgecolor = track.options.edgecolor 26 27 patch_options: dict[str, Any] = {"zorder": zorder} 28 patch_options["alpha"] = alpha 29 30 # Overlapping tracks should not cause the overlapped track to have their spines/ticks/ticklabels removed. 31 if track.pos != TrackPosition.Overlap: 32 spines = ( 33 ("right", "left", "top", "bottom") if hide_x else ("right", "left", "top") 34 ) 35 yticks = True 36 else: 37 yticks = False 38 spines = None 39 format_ax( 40 ax, 41 xticks=hide_x, 42 xticklabel_fontsize=track.options.fontsize, 43 yticks=yticks, 44 yticklabel_fontsize=track.options.fontsize, 45 spines=spines, 46 ) 47 48 ylim = ax.get_ylim() 49 height = ylim[1] - ylim[0] 50 51 patch_options["edgecolor"] = edgecolor 52 53 for row in track.data.iter_rows(named=True): 54 start = row["chrom_st"] 55 end = row["chrom_end"] 56 57 if row["name"] == "-" or not row["name"]: 58 labels = {} 59 else: 60 labels = {"label": row["name"]} 61 62 # Allow override. 63 if color: 64 patch_options["facecolor"] = color 65 elif "color" in row: 66 patch_options["facecolor"] = row["color"] 67 68 if track.options.shape == "rect": 69 rect = Rectangle( 70 (start, 0), 71 end + 1 - start, 72 height, 73 **labels, 74 **patch_options, 75 ) 76 ax.add_patch(rect) 77 elif track.options.shape == "tri": 78 midpt = ((end - start) / 2) + start 79 vertices = [ 80 (start, height), 81 (end, height), 82 # tip 83 (midpt, 0), 84 ] 85 ptch = Polygon( 86 vertices, 87 closed=True, 88 **labels, 89 **patch_options, 90 ) 91 ax.add_patch(ptch) 92 93 if border: 94 # Ensure border on top with larger zorder. 95 add_rect(ax, height, fill=False, zorder=zorder + 1.0) 96 97 # Draw legend. 98 if legend_ax and legend: 99 draw_uniq_entry_legend( 100 legend_ax, 101 track, 102 ref_ax=ax, 103 ncols=track.options.legend_ncols, 104 label_order=track.options.legend_label_order, 105 loc="center left", 106 alignment="left", 107 )
Draw label plot on axis with the given Track.
55def draw_self_ident( 56 ax: Axes, 57 track: Track, 58 *, 59 zorder: float = 1.0, 60 legend_ax: Axes | None = None, 61) -> None: 62 """ 63 Draw self identity plot on axis with the given `Track`. 64 """ 65 hide_x = track.options.hide_x 66 invert = track.options.invert 67 legend = track.options.legend 68 69 colors, verts = [], [] 70 spines = ("right", "left", "top", "bottom") if hide_x else ("right", "left", "top") 71 format_ax( 72 ax, 73 xticks=hide_x, 74 xticklabel_fontsize=track.options.fontsize, 75 yticks=True, 76 yticklabel_fontsize=track.options.fontsize, 77 spines=spines, 78 ) 79 80 if invert: 81 df_track = track.data.with_columns(y=-pl.col("y")) 82 else: 83 df_track = track.data 84 85 for _, df_diam in df_track.group_by(["group"]): 86 df_points = df_diam.select("x", "y") 87 color = df_diam["color"].first() 88 colors.append(color) 89 verts.append(df_points) 90 91 # https://stackoverflow.com/a/29000246 92 polys = PolyCollection(verts, zorder=zorder) 93 polys.set(array=None, facecolors=colors) 94 ax.add_collection(polys) 95 96 ymin, ymax = ( 97 df_track["y"].min(), 98 df_track["y"].max(), 99 ) 100 101 ax.set_ylim(ymin, ymax) 102 103 if legend_ax and legend: 104 draw_self_ident_hist(legend_ax, track, zorder=zorder)
Draw self identity plot on axis with the given Track.
12def draw_self_ident_hist(ax: Axes, track: Track, *, zorder: float = 1.0): 13 """ 14 Draw self identity histogram plot on axis with the given `Track`. 15 """ 16 legend_bins = track.options.legend_bins 17 legend_xmin = track.options.legend_xmin 18 legend_asp_ratio = track.options.legend_asp_ratio 19 colorscale = track.options.colorscale 20 assert isinstance(colorscale, dict), ( 21 f"Colorscale not a identity interval mapping for {track.title}" 22 ) 23 24 cmap = IntervalTree( 25 Interval(rng[0], rng[1], color) for rng, color in colorscale.items() 26 ) 27 cnts, values, bars = ax.hist( 28 track.data["percent_identity_by_events"], bins=legend_bins, zorder=zorder 29 ) 30 ax.set_xlim(legend_xmin, 100.0) 31 ax.minorticks_on() 32 ax.set_xlabel( 33 "Mean nucleotide identity\nbetween pairwise intervals", 34 fontsize=track.options.legend_title_fontsize, 35 ) 36 ax.set_ylabel( 37 "# of Intervals (thousands)", fontsize=track.options.legend_title_fontsize 38 ) 39 40 # Ensure that legend is only a portion of the total height. 41 # Otherwise, take up entire axis dim. 42 ax.set_box_aspect(legend_asp_ratio) 43 44 for _, value, bar in zip(cnts, values, bars): # type: ignore[arg-type] 45 # Make value a non-null interval 46 # ex. (1,1) -> (1, 1.000001) 47 color = cmap.overlap(value, value + 0.00001) 48 try: 49 color = next(iter(color)).data 50 except Exception: 51 color = None 52 bar.set_facecolor(color)
Draw self identity histogram plot on axis with the given Track.
9def draw_local_self_ident( 10 ax: Axes, 11 track: Track, 12 *, 13 zorder: float = 1.0, 14 legend_ax: Axes | None = None, 15) -> None: 16 """ 17 Draw local, self identity plot on axis with the given `Track`. 18 """ 19 if not track.options.legend_label_order: 20 track.options.legend_label_order = [ 21 f"{cs[0]}-{cs[1]}" for cs in track.options.colorscale.keys() 22 ] 23 draw_label(ax, track, zorder=zorder, legend_ax=legend_ax)
Draw local, self identity plot on axis with the given Track.
9def draw_bar( 10 ax: Axes, 11 track: Track, 12 *, 13 zorder: float = 1.0, 14 legend_ax: Axes | None = None, 15) -> None: 16 """ 17 Draw bar plot on axis with the given `Track`. 18 """ 19 hide_x = track.options.hide_x 20 color = track.options.color 21 alpha = track.options.alpha 22 legend = track.options.legend 23 label = track.options.label 24 25 if track.pos != TrackPosition.Overlap: 26 spines = ("right", "top") 27 else: 28 spines = None 29 30 format_ax( 31 ax, 32 xticks=hide_x, 33 xticklabel_fontsize=track.options.fontsize, 34 yticklabel_fontsize=track.options.fontsize, 35 spines=spines, 36 ) 37 38 plot_options = {"zorder": zorder, "alpha": alpha} 39 if color: 40 plot_options["color"] = color 41 elif "color" in track.data.columns: 42 plot_options["color"] = track.data["color"] 43 else: 44 plot_options["color"] = track.options.DEF_COLOR 45 46 # Add bar 47 ax.bar( 48 track.data["chrom_st"], 49 track.data["name"], 50 track.data["chrom_end"] - track.data["chrom_st"], 51 label=label, 52 **plot_options, 53 ) # type: ignore[arg-type] 54 # Trim plot to margins 55 ax.margins(x=0, y=0) 56 57 set_ylim(ax, track) 58 59 if legend_ax and legend: 60 draw_uniq_entry_legend( 61 legend_ax, 62 track, 63 ref_ax=ax, 64 ncols=track.options.legend_ncols, 65 loc="center left", 66 alignment="left", 67 )
Draw bar plot on axis with the given Track.
10def draw_line( 11 ax: Axes, 12 track: Track, 13 *, 14 zorder: float = 1.0, 15 legend_ax: Axes | None = None, 16) -> None: 17 """ 18 Draw line plot on axis with the given `Track`. 19 """ 20 hide_x = track.options.hide_x 21 color = track.options.color 22 alpha = track.options.alpha 23 legend = track.options.legend 24 label = track.options.label 25 linestyle = track.options.linestyle 26 linewidth = track.options.linewidth 27 marker = track.options.marker 28 markersize = track.options.markersize 29 30 if track.pos != TrackPosition.Overlap: 31 spines = ("right", "top") 32 else: 33 spines = None 34 35 format_ax( 36 ax, 37 xticks=hide_x, 38 xticklabel_fontsize=track.options.fontsize, 39 yticklabel_fontsize=track.options.fontsize, 40 spines=spines, 41 ) 42 43 plot_options = {"zorder": zorder, "alpha": alpha} 44 if color: 45 plot_options["color"] = color 46 elif "color" in track.data.columns: 47 plot_options["color"] = track.data["color"] 48 else: 49 plot_options["color"] = track.options.DEF_COLOR 50 51 if linestyle: 52 plot_options["linestyle"] = linestyle 53 if linewidth: 54 plot_options["linewidth"] = linewidth 55 56 # Fill between cannot add markers 57 if not track.options.fill: 58 plot_options["marker"] = marker 59 if markersize: 60 plot_options["markersize"] = markersize 61 62 if track.options.position == "midpoint": 63 df = track.data.with_columns( 64 chrom_st=pl.col("chrom_st") + (pl.col("chrom_end") - pl.col("chrom_st")) / 2 65 ) 66 else: 67 df = track.data 68 69 if track.options.log_scale: 70 ax.set_yscale("log") 71 72 # Add bar 73 if track.options.fill: 74 ax.fill_between( 75 df["chrom_st"], 76 df["name"], 77 0, 78 label=label, 79 **plot_options, 80 ) # type: ignore[arg-type] 81 else: 82 ax.plot( 83 df["chrom_st"], 84 df["name"], 85 label=label, 86 **plot_options, 87 ) # type: ignore[arg-type] 88 89 # Trim plot to margins 90 ax.margins(x=0, y=0) 91 92 set_ylim(ax, track) 93 94 if legend_ax and legend: 95 draw_uniq_entry_legend( 96 legend_ax, 97 track, 98 ref_ax=ax, 99 ncols=track.options.legend_ncols, 100 loc="center left", 101 alignment="left", 102 )
Draw line plot on axis with the given Track.
8def draw_strand( 9 ax: Axes, 10 track: Track, 11 *, 12 zorder: float = 1.0, 13 legend_ax: Axes | None = None, 14): 15 """ 16 Draw strand plot on axis with the given `Track`. 17 """ 18 hide_x = track.options.hide_x 19 fwd_color = ( 20 track.options.fwd_color if track.options.fwd_color else track.options.DEF_COLOR 21 ) 22 rev_color = ( 23 track.options.rev_color if track.options.rev_color else track.options.DEF_COLOR 24 ) 25 scale = track.options.scale 26 legend = track.options.legend 27 28 if track.pos != TrackPosition.Overlap: 29 spines = ( 30 ("right", "left", "top", "bottom") if hide_x else ("right", "left", "top") 31 ) 32 else: 33 spines = None 34 35 format_ax( 36 ax, 37 xticks=hide_x, 38 xticklabel_fontsize=track.options.fontsize, 39 yticks=True, 40 yticklabel_fontsize=track.options.fontsize, 41 spines=spines, 42 ) 43 44 ylim = ax.get_ylim() 45 height = ylim[1] - ylim[0] 46 47 for row in track.data.iter_rows(named=True): 48 # sample arrow 49 start = row["chrom_st"] 50 end = row["chrom_end"] 51 strand = row["strand"] 52 if strand == "-": 53 tmp_start = start 54 start = end 55 end = tmp_start 56 color = rev_color 57 else: 58 color = fwd_color 59 60 if track.options.use_item_rgb: 61 color = row["color"] 62 63 arrow = FancyArrowPatch( 64 (start, height * 0.5), 65 (end, height * 0.5), 66 mutation_scale=scale, 67 color=color, 68 clip_on=False, 69 zorder=zorder, 70 label=row["name"], 71 ) 72 ax.add_patch(arrow) 73 74 if legend_ax and legend: 75 draw_uniq_entry_legend( 76 legend_ax, 77 track, 78 ref_ax=ax, 79 ncols=track.options.legend_ncols, 80 loc="center", 81 )
Draw strand plot on axis with the given Track.
12def draw_legend( 13 ax: Axes, 14 axes: np.ndarray, 15 track: Track, 16 ref_track_col: int, 17) -> None: 18 """ 19 Draw legend plot on axis for the given `Track`. 20 21 # Args 22 * `ax` 23 * Axis to plot on. 24 * `axes` 25 * 2D `np.ndarray` of all axes to get reference axis. 26 * `track` 27 * Current `Track`. 28 * `track_col` 29 * Reference `Track` column. 30 31 # Returns 32 * None 33 """ 34 if isinstance(track.options.index, int): 35 ref_track_rows = [track.options.index] 36 elif isinstance(track.options.index, list): 37 ref_track_rows = track.options.index 38 else: 39 raise ValueError("Invalid type for reference legend indices.") 40 41 all_label_handles: dict[Any, Artist] = {} 42 for row in ref_track_rows: 43 try: 44 ref_track_ax: Axes = axes[row, ref_track_col] 45 except IndexError: 46 print(f"Reference axis index ({row}) doesn't exist.", sys.stderr) 47 continue 48 49 handles, labels = ref_track_ax.get_legend_handles_labels() 50 labels_handles: dict[Any, Artist] = dict(zip(labels, handles)) 51 all_label_handles = all_label_handles | labels_handles 52 53 # Provide custom order. 54 # Keeps all elements as opposed to draw_uniq_entry_legend 55 if track.options.legend_label_order: 56 legend_labels = set(all_label_handles.keys()) 57 new_all_label_handles = {} 58 for label in track.options.legend_label_order: 59 if not all_label_handles.get(label): 60 continue 61 new_all_label_handles[label] = all_label_handles[label] 62 63 remaining_labels = legend_labels.difference(new_all_label_handles.keys()) 64 for label in remaining_labels: 65 new_all_label_handles[label] = all_label_handles[label] 66 67 all_label_handles = new_all_label_handles 68 69 # Some code dup. 70 if not track.options.legend_title_only: 71 legend = ax.legend( 72 all_label_handles.values(), 73 all_label_handles.keys(), 74 ncols=track.options.legend_ncols if track.options.legend_ncols else 10, 75 # Set aspect ratio of handles so square. 76 handlelength=1.0, 77 handleheight=1.0, 78 frameon=False, 79 fontsize=track.options.legend_fontsize, 80 loc="center", 81 alignment="center", 82 ) 83 84 # Set patches edge color manually. 85 # Turns out get_legend_handles_labels will get all rect patches and setting linewidth will cause all patches to be black. 86 for ptch in legend.get_patches(): 87 ptch.set_linewidth(1.0) 88 ptch.set_edgecolor("black") 89 else: 90 legend = ax.legend([], [], frameon=False, loc="center left", alignment="left") 91 92 # Set legend title. 93 if track.options.legend_title: 94 legend.set_title(track.options.legend_title) 95 legend.get_title().set_fontsize(track.options.legend_title_fontsize) 96 97 format_ax( 98 ax, 99 grid=True, 100 xticks=True, 101 yticks=True, 102 spines=("right", "left", "top", "bottom"), 103 )
10def read_bed9(infile: str | TextIO, *, chrom: str | None = None) -> pl.DataFrame: 11 """ 12 Read a BED9 file with no header. 13 14 # Args 15 * `infile` 16 * Input file or IO stream. 17 * `chrom` 18 * Chromsome in `chrom` column to filter for. If contains coordinates, subset to those coordinates. 19 20 # Returns 21 * BED9 pl.DataFrame. 22 """ 23 skip_rows, number_cols = header_info(infile) 24 25 try: 26 df = pl.scan_csv( 27 infile, 28 separator="\t", 29 has_header=False, 30 skip_rows=skip_rows, 31 new_columns=BED9_COLS[0:number_cols], 32 ) 33 try: 34 chrom_no_coords, coords = chrom.rsplit(":", 1) 35 chrom_st, chrom_end = [int(elem) for elem in coords.split("-")] 36 except Exception: 37 chrom_no_coords = None 38 chrom_st, chrom_end = None, None 39 40 def expr_chrom_coords( 41 expr_no_coords: pl.Expr, expr_coords: pl.Expr, expr_otherwise: pl.Expr 42 ) -> pl.Expr: 43 return ( 44 pl.when(pl.col("chrom").eq(chrom_no_coords)) 45 .then(expr_no_coords) 46 .when(pl.col("chrom").eq(chrom)) 47 .then(expr_coords) 48 .otherwise(expr_otherwise) 49 ) 50 51 # Chrom coordinates can be one of three states: 52 # 1. chr1:0-10:0-5 53 # 2. chr1:0-10 54 # 3. chr1 55 # We assume if an exact match for chrom_no_coords is found (1), the user wants to trim to some coordinates. 56 # Coordinate are right split once. 57 if chrom_no_coords and chrom_st and chrom_end: 58 df_filtered = ( 59 df.filter( 60 expr_chrom_coords( 61 pl.col("chrom") == chrom_no_coords, 62 pl.col("chrom") == chrom, 63 True, 64 ) 65 ) 66 .with_columns( 67 chrom_st=expr_chrom_coords( 68 pl.col("chrom_st").clip(chrom_st, chrom_end), 69 pl.col("chrom_st"), 70 pl.col("chrom_st"), 71 ), 72 chrom_end=expr_chrom_coords( 73 pl.col("chrom_end").clip(chrom_st, chrom_end), 74 pl.col("chrom_end"), 75 pl.col("chrom_end"), 76 ), 77 ) 78 # Remove null intervals created by clipping to boundaries 79 .filter( 80 expr_chrom_coords( 81 ~( 82 ( 83 pl.col("chrom_st").eq(chrom_st) 84 & pl.col("chrom_st").eq(chrom_end) 85 ) 86 | ( 87 pl.col("chrom_end").eq(chrom_st) 88 & pl.col("chrom_end").eq(chrom_end) 89 ) 90 ), 91 True, 92 True, 93 ) 94 ) 95 .collect() 96 ) 97 elif chrom: 98 df_filtered = df.filter(pl.col("chrom") == chrom).collect() 99 else: 100 df_filtered = df.collect() 101 102 df_adj = adj_by_ctg_coords(df_filtered, "chrom").sort(by="chrom_st") 103 except pl.exceptions.NoDataError: 104 df_adj = pl.DataFrame(schema=BED9_COLS) 105 106 if "item_rgb" not in df_adj.columns: 107 df_adj = df_adj.with_columns(item_rgb=pl.lit("0,0,0")) 108 if "name" not in df_adj.columns: 109 df_adj = df_adj.with_columns(name=pl.lit("-")) 110 111 return df_adj
Read a BED9 file with no header.
Args
infile- Input file or IO stream.
chrom- Chromsome in
chromcolumn to filter for. If contains coordinates, subset to those coordinates.
- Chromsome in
Returns
- BED9 pl.DataFrame.
14def read_bed_hor( 15 infile: str | TextIO, 16 *, 17 chrom: str | None = None, 18 live_only: bool = True, 19 mer_size: int = HORTrackSettings.mer_size, 20 mer_filter: int = HORTrackSettings.mer_filter, 21 hor_filter: int | None = None, 22 sort_by: str = "mer", 23 sort_order: str = HORTrackSettings.sort_order, 24 sort_fill_missing: str | None = HORTrackSettings.split_fill_missing, 25 sort_order_only: bool = False, 26 color_map_file: str | None = None, 27 use_item_rgb: bool = HORTrackSettings.use_item_rgb, 28) -> pl.DataFrame: 29 """ 30 Read a HOR BED9 file with no header. 31 32 # Args 33 * `infile` 34 * Input file or IO stream. 35 * `chrom` 36 * Chromsome in `chrom` column to filter for. 37 * `live_only` 38 * Filter for only live data. 39 * Contains `L` in `name` column. 40 * `mer_size` 41 * Monomer size to calculate monomer number. 42 * `mer_filter` 43 * Filter for HORs with at least this many monomers. 44 * `hor_filter` 45 * Filter for HORs that occur at least this many times. 46 * `color_map_file` 47 * Convenience color map file for `mer` or `hor`. 48 * Two-column TSV file with no header. 49 * If `None`, use default color map. 50 * `sort_by` 51 * Sort `pl.DataFrame` by `mer`, `hor`, or `hor_count`. 52 * Can be a path to a list of `mer` or `hor` names 53 * `sort_order` 54 * Sort in ascending or descending order. 55 * `sort_fill_missing` 56 * Fill in missing elements in defined sort order with this color. 57 * `sort_order_only` 58 * Convenience switch to keep only elements in defined sort order. 59 * `use_item_rgb` 60 * Use `item_rgb` column or generate random colors. 61 62 # Returns 63 * HOR `pl.DataFrame` 64 """ 65 df = read_bed9(infile, chrom=chrom) 66 67 if df.is_empty(): 68 return pl.DataFrame(schema=[*BED9_COLS, "mer", "length", "color", "hor_count"]) 69 70 df = ( 71 df.lazy() 72 .with_columns( 73 length=pl.col("chrom_end") - pl.col("chrom_st"), 74 ) 75 .with_columns( 76 mer=(pl.col("length") / mer_size).round().cast(pl.UInt32).clip(1, 100) 77 ) 78 .filter( 79 pl.when(live_only).then(pl.col("name").str.contains("L")).otherwise(True) 80 & (pl.col("mer") >= mer_filter) 81 ) 82 .collect() 83 ) 84 # Read color map. 85 if color_map_file: 86 color_map: dict[str, str] = {} 87 with open(color_map_file, "rt") as fh: 88 for line in fh.readlines(): 89 try: 90 name, color = line.strip().split() 91 except Exception: 92 logging.error(f"Invalid color map. ({line})") 93 continue 94 color_map[name] = color 95 else: 96 color_map = MONOMER_COLORS 97 98 df = map_value_colors( 99 df, 100 map_col="mer", 101 map_values=MONOMER_COLORS, 102 use_item_rgb=use_item_rgb, 103 ) 104 df = df.join(df.get_column("name").value_counts(name="hor_count"), on="name") 105 106 if hor_filter: 107 df = df.filter(pl.col("hor_count") >= hor_filter) 108 109 if os.path.exists(sort_order): 110 with open(sort_order, "rt") as fh: 111 defined_sort_order = [] 112 for line in fh: 113 line = line.strip() 114 defined_sort_order.append(int(line) if sort_by == "mer" else line) 115 else: 116 defined_sort_order = None 117 118 if sort_by == "mer": 119 sort_col = "mer" 120 elif sort_by == "name" and defined_sort_order: 121 sort_col = "name" 122 else: 123 sort_col = "hor_count" 124 125 if defined_sort_order: 126 # Add missing elems in df not in sort order so all elements covered. 127 all_elems = [ 128 *defined_sort_order, 129 *set(df[sort_col]).difference(defined_sort_order), 130 ] 131 # Missing elements in sort order not in df 132 missing_elems = set(defined_sort_order).difference(df[sort_col]) 133 134 # Fill in missing. 135 if sort_fill_missing and missing_elems: 136 row_template = df.row(0, named=True) 137 min_st, max_end = df["chrom_st"].min(), df["chrom_end"].max() 138 df_missing_element_rows = pl.DataFrame( 139 [ 140 { 141 **row_template, 142 "chrom_st": min_st, 143 "chrom_end": max_end, 144 "strand": ".", 145 "thick_st": min_st, 146 "thick_end": max_end, 147 "name": elem, 148 "mer": 0, 149 "length": 0, 150 "hor_count": 0, 151 "item_rgb": sort_fill_missing, 152 "color": sort_fill_missing, 153 } 154 for elem in missing_elems 155 ], 156 schema=df.schema, 157 ) 158 159 df = pl.concat([df, df_missing_element_rows]) 160 # Only take elements in sort order. 161 if sort_order_only: 162 df = df.filter(pl.col(sort_col).is_in(defined_sort_order)) 163 all_elems = defined_sort_order 164 165 df = df.cast({sort_col: pl.Enum(all_elems)}).sort(by=sort_col) 166 else: 167 df = df.sort(sort_col, descending=sort_order == HORTrackSettings.sort_order) 168 169 return df
Read a HOR BED9 file with no header.
Args
infile- Input file or IO stream.
chrom- Chromsome in
chromcolumn to filter for.
- Chromsome in
live_only- Filter for only live data.
- Contains
Linnamecolumn.
mer_size- Monomer size to calculate monomer number.
mer_filter- Filter for HORs with at least this many monomers.
hor_filter- Filter for HORs that occur at least this many times.
color_map_file- Convenience color map file for
merorhor. - Two-column TSV file with no header.
- If
None, use default color map.
- Convenience color map file for
sort_by- Sort
pl.DataFramebymer,hor, orhor_count. - Can be a path to a list of
merorhornames
- Sort
sort_order- Sort in ascending or descending order.
sort_fill_missing- Fill in missing elements in defined sort order with this color.
sort_order_only- Convenience switch to keep only elements in defined sort order.
use_item_rgb- Use
item_rgbcolumn or generate random colors.
- Use
Returns
- HOR
pl.DataFrame
198def read_bed_identity( 199 infile: str | TextIO, 200 *, 201 chrom: str | None = None, 202 mode: str = "2D", 203 colorscale: Colorscale | str | None = None, 204 band_size: int = LocalSelfIdentTrackSettings.band_size, 205 ignore_band_size=LocalSelfIdentTrackSettings.ignore_band_size, 206) -> tuple[pl.DataFrame, Colorscale]: 207 """ 208 Read a self, sequence identity BED file generate by `ModDotPlot`. 209 210 Requires the following columns 211 * `query,query_st,query_end,ref,ref_st,ref_end,percent_identity_by_events` 212 213 # Args 214 * `infile` 215 * File or IO stream. 216 * `chrom` 217 * Chromosome name in `query` column to filter for. 218 * `mode` 219 * 1D or 2D self-identity. 220 * `band_size` 221 * Number of windows to calculate average sequence identity over. Only applicable if mode is 1D. 222 * `ignore_band_size` 223 * Number of windows ignored along self-identity diagonal. Only applicable if mode is 1D. 224 225 # Returns 226 * Coordinates of colored polygons in 2D space. 227 """ 228 df = read_bedpe(infile=infile, chrom=chrom) 229 230 # Check mode. Set by dev not user. 231 mode = Dim(mode) 232 233 # Build expr to filter range of colors. 234 color_expr = None 235 rng_expr = None 236 ident_colorscale = read_ident_colorscale(colorscale) 237 for rng, color in ident_colorscale.items(): 238 if not isinstance(color_expr, pl.Expr): 239 color_expr = pl.when( 240 pl.col("percent_identity_by_events").is_between(rng[0], rng[1]) 241 ).then(pl.lit(color)) # type: ignore[assignment] 242 rng_expr = pl.when( 243 pl.col("percent_identity_by_events").is_between(rng[0], rng[1]) 244 ).then(pl.lit(f"{rng[0]}-{rng[1]}")) # type: ignore[assignment] 245 else: 246 color_expr = color_expr.when( 247 pl.col("percent_identity_by_events").is_between(rng[0], rng[1]) 248 ).then(pl.lit(color)) # type: ignore[assignment] 249 rng_expr = rng_expr.when( 250 pl.col("percent_identity_by_events").is_between(rng[0], rng[1]) 251 ).then(pl.lit(f"{rng[0]}-{rng[1]}")) # type: ignore[assignment] 252 253 if isinstance(color_expr, pl.Expr): 254 color_expr = color_expr.otherwise(None) # type: ignore[assignment] 255 else: 256 color_expr = pl.lit(None) # type: ignore[assignment] 257 if isinstance(rng_expr, pl.Expr): 258 rng_expr = rng_expr.otherwise(None) # type: ignore[assignment] 259 else: 260 rng_expr = pl.lit(None) # type: ignore[assignment] 261 262 if mode == Dim.ONE: 263 df_window = ( 264 (df["query_end"] - df["query_st"]) 265 .value_counts(sort=True) 266 .rename({"query_end": "window"}) 267 ) 268 if df_window.shape[0] > 1: 269 logging.warning(f"Multiple windows detected. Taking largest.\n{df_window}") 270 window = df_window.row(0, named=True)["window"] + 1 271 df_local_ident = pl.DataFrame( 272 convert_2D_to_1D_ident(df.iter_rows(), window, band_size, ignore_band_size), 273 schema=[ 274 "chrom_st", 275 "chrom_end", 276 "percent_identity_by_events", 277 ], 278 orient="row", 279 ) 280 query = df["query"][0] 281 df_res = ( 282 df_local_ident.lazy() 283 .with_columns( 284 chrom=pl.lit(query), 285 color=color_expr, 286 name=rng_expr, 287 score=pl.col("percent_identity_by_events"), 288 strand=pl.lit("."), 289 thick_st=pl.col("chrom_st"), 290 thick_end=pl.col("chrom_end"), 291 item_rgb=pl.lit("0,0,0"), 292 ) 293 .select(*BED9_COLS, "color") 294 .collect() 295 ) 296 else: 297 tri_side = math.sqrt(2) / 2 298 df_res = ( 299 df.lazy() 300 .with_columns(color=color_expr) 301 # Get window size. 302 .with_columns( 303 window=(pl.col("query_end") - pl.col("query_st")).max().over("query") 304 ) 305 .with_columns( 306 first_pos=pl.col("query_st") // pl.col("window"), 307 second_pos=pl.col("ref_st") // pl.col("window"), 308 ) 309 # x y coords of diamond 310 .with_columns( 311 x=pl.col("first_pos") + pl.col("second_pos"), 312 y=-pl.col("first_pos") + pl.col("second_pos"), 313 ) 314 .with_columns( 315 scale=(pl.col("query_st").max() / pl.col("x").max()).over("query"), 316 group=pl.int_range(pl.len()).over("query"), 317 ) 318 .with_columns( 319 window=pl.col("window") / pl.col("scale"), 320 ) 321 # Rather than generate new dfs. Add new x,y as arrays per row. 322 .with_columns( 323 new_x=[tri_side, 0.0, -tri_side, 0.0], 324 new_y=[0.0, tri_side, 0.0, -tri_side], 325 ) 326 # Rescale x and y. 327 .with_columns( 328 ((pl.col("new_x") * pl.col("window")) + pl.col("x")) * pl.col("scale"), 329 ((pl.col("new_y") * pl.col("window")) + pl.col("y")) * pl.col("window"), 330 ) 331 .select( 332 "query", 333 "new_x", 334 "new_y", 335 "color", 336 "group", 337 "percent_identity_by_events", 338 ) 339 # arr to new rows 340 .explode("new_x", "new_y") 341 # Rename to filter later on. 342 .rename({"query": "chrom", "new_x": "x", "new_y": "y"}) 343 .collect() 344 ) 345 return df_res, ident_colorscale
Read a self, sequence identity BED file generate by ModDotPlot.
Requires the following columns
query,query_st,query_end,ref,ref_st,ref_end,percent_identity_by_events
Args
infile- File or IO stream.
chrom- Chromosome name in
querycolumn to filter for.
- Chromosome name in
mode- 1D or 2D self-identity.
band_size- Number of windows to calculate average sequence identity over. Only applicable if mode is 1D.
ignore_band_size- Number of windows ignored along self-identity diagonal. Only applicable if mode is 1D.
Returns
- Coordinates of colored polygons in 2D space.
9def read_bed_label(infile: str | TextIO, *, chrom: str | None = None) -> pl.DataFrame: 10 """ 11 Read a BED9 file with no header. 12 * Labels are ordered by length. 13 14 # Args 15 * `infile` 16 * Input file or IO stream. 17 * `chrom` 18 * Chromsome in `chrom` column to filter for. 19 20 # Returns 21 * BED9 pl.DataFrame. 22 """ 23 df_track = read_bed9(infile, chrom=chrom) 24 25 # Order facets by descending length. This prevents larger annotations from blocking others. 26 fct_name_order = ( 27 df_track.group_by(["name"]) 28 .agg(len=(pl.col("chrom_end") - pl.col("chrom_st")).sum()) 29 .sort(by="len", descending=True) 30 .get_column("name") 31 ) 32 return df_track.cast({"name": pl.Enum(fct_name_order)})
Read a BED9 file with no header.
- Labels are ordered by length.
Args
infile- Input file or IO stream.
chrom- Chromsome in
chromcolumn to filter for.
- Chromsome in
Returns
- BED9 pl.DataFrame.
86def read_track( 87 track: dict[str, Any], *, chrom: str | None = None 88) -> Generator[Track, None, None]: 89 prop = track.get("proportion", 0.0) 90 title = track.get("title") 91 pos = track.get("position") 92 opt = track.get("type") 93 path: str | None = track.get("path") 94 options: dict[str, Any] = track.get("options", {}) 95 96 try: 97 track_pos = TrackPosition(pos) # type: ignore[arg-type] 98 except ValueError: 99 logging.error(f"Invalid plot position ({pos}) for {path}. Skipping.") 100 return None 101 try: 102 track_opt = TrackType(opt) # type: ignore[arg-type] 103 except ValueError: 104 logging.error(f"Invalid plot option ({opt}) for {path}. Skipping.") 105 return None 106 107 track_options: TrackSettings 108 if track_opt == TrackType.Position: 109 track_options = PositionTrackSettings(**options) 110 track_options.hide_x = False 111 yield Track(title, track_pos, track_opt, prop, pl.DataFrame(), track_options) 112 return None 113 elif track_opt == TrackType.Legend: 114 track_options = LegendTrackSettings(**options) 115 yield Track(title, track_pos, track_opt, prop, pl.DataFrame(), track_options) 116 return None 117 elif track_opt == TrackType.Spacer: 118 track_options = SpacerTrackSettings(**options) 119 yield Track(title, track_pos, track_opt, prop, pl.DataFrame(), track_options) 120 return None 121 122 if not path: 123 raise ValueError("Path to data required.") 124 125 if not os.path.exists(path): 126 raise FileNotFoundError(f"Data does not exist for track ({track})") 127 128 if track_opt == TrackType.HORSplit: 129 df_track = read_bed_hor_from_settings(path, options, chrom) 130 if df_track.is_empty(): 131 logging.error( 132 f"Empty file or chrom not found for {track_opt} and {path}. Skipping" 133 ) 134 return None 135 if options.get("mode", HORTrackSettings.mode) == "hor": 136 split_colname = "name" 137 else: 138 split_colname = "mer" 139 split_prop = options.get("split_prop", HORTrackSettings.split_prop) 140 yield from split_hor_track( 141 df_track, 142 track_pos, 143 track_opt, 144 title, 145 prop, 146 split_colname, 147 split_prop, 148 options, 149 chrom=chrom, 150 ) 151 return None 152 153 elif track_opt == TrackType.HOR: 154 df_track = read_bed_hor_from_settings(path, options, chrom) 155 track_options = HORTrackSettings(**options) 156 # Update legend title. 157 if track_options.legend_title: 158 track_options.legend_title = track_options.legend_title.format(chrom=chrom) 159 160 yield Track(title, track_pos, track_opt, prop, df_track, track_options) 161 return None 162 163 if track_opt == TrackType.HOROrt: 164 live_only = options.get("live_only", HOROrtTrackSettings.live_only) 165 mer_filter = options.get("mer_filter", HOROrtTrackSettings.mer_filter) 166 hor_length_kwargs = { 167 "output_strand": True, 168 "allow_nonlive": not live_only, 169 } 170 # HOR array length args are prefixed with `arr_opt_` 171 for opt, value in options.items(): 172 if opt.startswith("arr_opt_"): 173 k = opt.replace("arr_opt_", "") 174 hor_length_kwargs[k] = value 175 176 df_hor = read_bed_hor( 177 path, 178 chrom=chrom, 179 live_only=live_only, 180 mer_filter=mer_filter, 181 ) 182 try: 183 _, df_track = hor_array_length(df_hor, **hor_length_kwargs) 184 except ValueError: 185 logging.error(f"Failed to calculate HOR array length for {path}.") 186 df_track = pl.DataFrame( 187 schema=[ 188 "chrom", 189 "chrom_st", 190 "chrom_end", 191 "name", 192 "score", 193 "prop", 194 "strand", 195 ] 196 ) 197 track_options = HOROrtTrackSettings(**options) 198 elif track_opt == TrackType.Strand: 199 use_item_rgb = options.get("use_item_rgb", StrandTrackSettings.use_item_rgb) 200 df_track = read_bed9(path, chrom=chrom) 201 df_track = map_value_colors(df_track, use_item_rgb=use_item_rgb) 202 track_options = StrandTrackSettings(**options) 203 elif track_opt == TrackType.SelfIdent: 204 df_track, colorscale = read_bed_identity( 205 path, chrom=chrom, colorscale=options.get("colorscale") 206 ) 207 # Save colorscale 208 options["colorscale"] = colorscale 209 210 track_options = SelfIdentTrackSettings(**options) 211 elif track_opt == TrackType.LocalSelfIdent: 212 band_size = options.get("band_size", LocalSelfIdentTrackSettings.band_size) 213 ignore_band_size = options.get( 214 "ignore_band_size", LocalSelfIdentTrackSettings.ignore_band_size 215 ) 216 df_track, colorscale = read_bed_identity( 217 path, 218 chrom=chrom, 219 mode="1D", 220 band_size=band_size, 221 ignore_band_size=ignore_band_size, 222 colorscale=options.get("colorscale"), 223 ) 224 # Save colorscale 225 options["colorscale"] = colorscale 226 227 track_options = LocalSelfIdentTrackSettings(**options) 228 elif track_opt == TrackType.Bar: 229 df_track = read_bed9(path, chrom=chrom) 230 track_options = BarTrackSettings(**options) 231 elif track_opt == TrackType.Line: 232 df_track = read_bed9(path, chrom=chrom) 233 track_options = LineTrackSettings(**options) 234 else: 235 use_item_rgb = options.get("use_item_rgb", LabelTrackSettings.use_item_rgb) 236 df_track = read_bed_label(path, chrom=chrom) 237 df_track = map_value_colors( 238 df_track, 239 map_col="name", 240 use_item_rgb=use_item_rgb, 241 ) 242 track_options = LabelTrackSettings(**options) 243 244 df_track = map_value_colors(df_track) 245 # Update legend title. 246 if track_options.legend_title: 247 track_options.legend_title = track_options.legend_title.format(chrom=chrom) 248 249 yield Track(title, track_pos, track_opt, prop, df_track, track_options)
252def read_tracks( 253 input_track: BinaryIO, *, chrom: str | None = None 254) -> tuple[TrackList, PlotSettings]: 255 """ 256 Read a `TOML` or `YAML` file of tracks to plot optionally filtering for a chrom name. 257 258 Expected to have two items: 259 * `[settings]` 260 * See `cenplot.PlotSettings` 261 * `[[tracks]]` 262 * See one of the `cenplot.TrackSettings` for more details. 263 264 Example: 265 ```toml 266 [settings] 267 format = "png" 268 transparent = true 269 dim = [16.0, 8.0] 270 dpi = 600 271 ``` 272 273 ```yaml 274 settings: 275 format: "png" 276 transparent: true 277 dim: [16.0, 8.0] 278 dpi: 600 279 ``` 280 281 # Args: 282 * input_track: 283 * Input track `TOML` or `YAML` file. 284 * chrom: 285 * Chromosome name in 1st column (`chrom`) to filter for. 286 * ex. `chr4` 287 288 # Returns: 289 * List of tracks w/contained chroms and plot settings. 290 """ 291 all_tracks = [] 292 chroms: set[str] = set() 293 # Reset file position. 294 input_track.seek(0) 295 # Try TOML 296 try: 297 dict_settings = tomllib.load(input_track) 298 except Exception: 299 input_track.seek(0) 300 # Then YAML 301 try: 302 dict_settings = yaml.safe_load(input_track) 303 except Exception: 304 raise TypeError("Invalid file type for settings.") 305 306 settings: dict[str, Any] = dict_settings.get("settings", {}) 307 if settings.get("dim"): 308 settings["dim"] = tuple(settings["dim"]) 309 310 for track_info in dict_settings.get("tracks", []): 311 for track in read_track(track_info, chrom=chrom): 312 all_tracks.append(track) 313 # Tracks legend and position have no data. 314 if track.data.is_empty(): 315 continue 316 chroms.update(track.data["chrom"]) 317 tracklist = TrackList(all_tracks, chroms) 318 319 _, min_st_pos = get_min_max_track(all_tracks, typ="min") 320 _, max_end_pos = get_min_max_track(all_tracks, typ="max", default_col="chrom_end") 321 if settings.get("xlim"): 322 settings["xlim"] = tuple(settings["xlim"]) 323 else: 324 settings["xlim"] = (min_st_pos, max_end_pos) 325 326 plot_settings = PlotSettings(**settings) 327 return tracklist, plot_settings
Read a TOML or YAML file of tracks to plot optionally filtering for a chrom name.
Expected to have two items:
[settings][[tracks]]- See one of the
cenplot.TrackSettingsfor more details.
- See one of the
Example:
[settings]
format = "png"
transparent = true
dim = [16.0, 8.0]
dpi = 600
settings:
format: "png"
transparent: true
dim: [16.0, 8.0]
dpi: 600
Args:
- input_track:
- Input track
TOMLorYAMLfile.
- Input track
- chrom:
- Chromosome name in 1st column (
chrom) to filter for. - ex.
chr4
- Chromosome name in 1st column (
Returns:
- List of tracks w/contained chroms and plot settings.
186@dataclass 187class Track: 188 """ 189 A centromere track. 190 """ 191 192 title: str | None 193 """ 194 Title of track. 195 * ex. "{chrom}" 196 * ex. "HOR monomers" 197 """ 198 pos: TrackPosition 199 """ 200 Track position. 201 """ 202 opt: TrackType 203 """ 204 Track option. 205 """ 206 prop: float 207 """ 208 Proportion of track in final figure. 209 """ 210 data: pl.DataFrame 211 """ 212 Track data. 213 """ 214 options: TrackSettings # type: ignore 215 """ 216 Plot settings. 217 """
A centromere track.
Plot settings.
28class TrackType(StrEnum): 29 """ 30 Track options. 31 * Input track data is expected to be headerless. 32 """ 33 34 HOR = auto() 35 """ 36 An alpha-satellite higher order repeat (HOR) track with HORs by monomer number overlapping. 37 38 Expected format: 39 * [`BED9`](https://genome.ucsc.edu/FAQ/FAQformat.html#format1) 40 * `name` as HOR variant 41 * ex. `S4CYH1L.44-1` 42 """ 43 HORSplit = auto() 44 """ 45 A split alpha-satellite higher order repeat (HOR) track with each type of HOR as a single track. 46 * `mer` or the number of monomers within the HOR. 47 * `hor` or HOR variant. 48 49 Expected format: 50 * [`BED9`](https://genome.ucsc.edu/FAQ/FAQformat.html#format1) 51 * `name` as HOR variant 52 * ex. `S4CYH1L.44-1` 53 """ 54 HOROrt = auto() 55 """ 56 An alpha-satellite higher order repeat (HOR) orientation track. 57 * This is calculate with default settings via the [`censtats`](https://github.com/logsdon-lab/CenStats) library. 58 59 Expected format: 60 * [`BED9`](https://genome.ucsc.edu/FAQ/FAQformat.html#format1) 61 * `name` as HOR variant 62 * ex. `S4CYH1L.44-1` 63 * `strand` as `+` or `-` 64 """ 65 Label = auto() 66 """ 67 A label track. Elements in the `name` column are displayed as colored rectangles. 68 69 Expected format: 70 * [`BED4-9`](https://genome.ucsc.edu/FAQ/FAQformat.html#format1) 71 * `name` as any string value. 72 """ 73 Bar = auto() 74 """ 75 A bar plot track. Elements in the `name` column are displayed as bars. 76 77 Expected format: 78 * `BED9` 79 * `name` as any numeric value. 80 """ 81 82 Line = auto() 83 """ 84 A line plot track. 85 86 Expected format: 87 * `BED9` 88 * `name` as any numeric value. 89 """ 90 91 SelfIdent = auto() 92 """ 93 A self, sequence identity heatmap track displayed as a triangle. 94 * Similar to plots from [`ModDotPlot`](https://github.com/marbl/ModDotPlot) 95 96 Expected format: 97 * `BEDPE*` 98 * Paired identity bedfile produced by `ModDotPlot` without a header. 99 100 |query|query_st|query_end|reference|reference_st|reference_end|percent_identity_by_events| 101 |-|-|-|-|-|-|-| 102 |x|1|5000|x|1|5000|100.0| 103 104 """ 105 LocalSelfIdent = auto() 106 """ 107 A self, sequence identity track showing local identity. 108 * Derived from [`ModDotPlot`](https://github.com/marbl/ModDotPlot) 109 110 Expected format: 111 * `BEDPE*` 112 * Paired identity bedfile produced by `ModDotPlot` without a header. 113 114 |query|query_st|query_end|reference|reference_st|reference_end|percent_identity_by_events| 115 |-|-|-|-|-|-|-| 116 |x|1|5000|x|1|5000|100.0| 117 """ 118 119 Strand = auto() 120 """ 121 Strand track. 122 123 Expected format: 124 * `BED9` 125 * `strand` as either `+` or `-` 126 """ 127 128 Position = auto() 129 """ 130 Position track. 131 * Displays the x-axis position as well as a label. 132 133 Expected format: 134 * None 135 """ 136 137 Legend = auto() 138 """ 139 Legend track. Displays the legend of a specified track. 140 * NOTE: This does not work with `TrackType.HORSplit` 141 142 Expected format: 143 * None 144 """ 145 146 Spacer = auto() 147 """ 148 Spacer track. Empty space. 149 150 Expected format: 151 * None 152 """ 153 154 def settings(self) -> TrackSettings: 155 """ 156 Get settings for track type. 157 """ 158 if self == TrackType.Bar: 159 return BarTrackSettings() 160 elif self == TrackType.HOR: 161 return HORTrackSettings() 162 elif self == TrackType.HOROrt: 163 return HOROrtTrackSettings() 164 elif self == TrackType.HORSplit: 165 return HORTrackSettings() 166 elif self == TrackType.Label: 167 return LabelTrackSettings() 168 elif self == TrackType.Legend: 169 return LegendTrackSettings() 170 elif self == TrackType.Line: 171 return LineTrackSettings() 172 elif self == TrackType.LocalSelfIdent: 173 return LocalSelfIdentTrackSettings() 174 elif self == TrackType.SelfIdent: 175 return SelfIdentTrackSettings() 176 elif self == TrackType.Position: 177 return PositionTrackSettings() 178 elif self == TrackType.Spacer: 179 return SpacerTrackSettings() 180 elif self == TrackType.Strand: 181 return StrandTrackSettings() 182 else: 183 raise ValueError(f"No settings provided for track type. {self}")
Track options.
- Input track data is expected to be headerless.
A self, sequence identity heatmap track displayed as a triangle.
- Similar to plots from
ModDotPlot
Expected format:
BEDPE*- Paired identity bedfile produced by
ModDotPlotwithout a header.
- Paired identity bedfile produced by
| query | query_st | query_end | reference | reference_st | reference_end | percent_identity_by_events |
|---|---|---|---|---|---|---|
| x | 1 | 5000 | x | 1 | 5000 | 100.0 |
A self, sequence identity track showing local identity.
- Derived from
ModDotPlot
Expected format:
BEDPE*- Paired identity bedfile produced by
ModDotPlotwithout a header.
- Paired identity bedfile produced by
| query | query_st | query_end | reference | reference_st | reference_end | percent_identity_by_events |
|---|---|---|---|---|---|---|
| x | 1 | 5000 | x | 1 | 5000 | 100.0 |
Position track.
- Displays the x-axis position as well as a label.
Expected format:
- None
Legend track. Displays the legend of a specified track.
- NOTE: This does not work with
TrackType.HORSplit
Expected format:
- None
154 def settings(self) -> TrackSettings: 155 """ 156 Get settings for track type. 157 """ 158 if self == TrackType.Bar: 159 return BarTrackSettings() 160 elif self == TrackType.HOR: 161 return HORTrackSettings() 162 elif self == TrackType.HOROrt: 163 return HOROrtTrackSettings() 164 elif self == TrackType.HORSplit: 165 return HORTrackSettings() 166 elif self == TrackType.Label: 167 return LabelTrackSettings() 168 elif self == TrackType.Legend: 169 return LegendTrackSettings() 170 elif self == TrackType.Line: 171 return LineTrackSettings() 172 elif self == TrackType.LocalSelfIdent: 173 return LocalSelfIdentTrackSettings() 174 elif self == TrackType.SelfIdent: 175 return SelfIdentTrackSettings() 176 elif self == TrackType.Position: 177 return PositionTrackSettings() 178 elif self == TrackType.Spacer: 179 return SpacerTrackSettings() 180 elif self == TrackType.Strand: 181 return StrandTrackSettings() 182 else: 183 raise ValueError(f"No settings provided for track type. {self}")
Get settings for track type.
220class TrackList(NamedTuple): 221 """ 222 Track list. 223 """ 224 225 tracks: list[Track] 226 """ 227 Tracks. 228 """ 229 chroms: set[str] 230 """ 231 Chromosomes found with `tracks`. 232 """
Track list.
9@dataclass 10class PlotSettings: 11 """ 12 Plot settings for a single plot. 13 """ 14 15 title: str | None = None 16 """ 17 Figure title. 18 19 Can use "{chrom}" to replace with chrom name. 20 """ 21 22 title_x: float | None = 0.02 23 """ 24 Figure title x position. 25 """ 26 27 title_y: float | None = None 28 """ 29 Figure title y position. 30 """ 31 32 title_fontsize: float | str = "xx-large" 33 """ 34 Figure title fontsize. 35 """ 36 37 title_horizontalalignment: str = "left" 38 """ 39 Figure title position. 40 """ 41 42 format: list[OutputFormat] | OutputFormat = "png" 43 """ 44 Output format(s). Either `"pdf"`, `"png"`, or `"svg"`. 45 """ 46 transparent: bool = True 47 """ 48 Output a transparent image. 49 """ 50 dim: tuple[float, float] = (20.0, 12.0) 51 """ 52 The dimensions of each plot. Format: `(width, height)` 53 """ 54 dpi: int = 600 55 """ 56 Set the plot DPI per plot. 57 """ 58 layout: str = "tight" 59 """ 60 Layout engine option for matplotlib. See https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.figure.html#matplotlib.pyplot.figure. 61 """ 62 legend_pos: LegendPosition = LegendPosition.Right 63 """ 64 Legend position as `LegendPosition`. Either `LegendPosition.Right` or `LegendPosition.Left`. 65 """ 66 legend_prop: float = 0.2 67 """ 68 Legend proportion of plot. 69 """ 70 axis_h_pad: float = 0.2 71 """ 72 Apply a height padding to each axis. 73 """ 74 xlim: tuple[int, int] | None = None 75 """ 76 Set x-axis limit across all plots. 77 * `None` - Use the min and max position across all tracks. 78 * `tuple[float, float]` - Use provided coordinates as min and max position. 79 """
Plot settings for a single plot.
Output format(s). Either "pdf", "png", or "svg".
Layout engine option for matplotlib. See https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.figure.html#matplotlib.pyplot.figure.
Legend position as LegendPosition. Either LegendPosition.Right or LegendPosition.Left.
71@dataclass 72class SelfIdentTrackSettings(DefaultTrackSettings): 73 """ 74 Self-identity heatmap triangle plot options. 75 """ 76 77 invert: bool = True 78 """ 79 Invert the self identity triangle. 80 """ 81 legend_bins: int = 300 82 """ 83 Number of bins for `perc_identity_by_events` in the legend. 84 """ 85 legend_xmin: float = 70.0 86 """ 87 Legend x-min coordinate. Used to constrain x-axis limits. 88 """ 89 legend_asp_ratio: float | None = 1.0 90 """ 91 Aspect ratio of legend. If `None`, takes up entire axis. 92 """ 93 colorscale: Colorscale | str | None = None 94 """ 95 Colorscale for identity as TSV file. 96 * Format: `[start, end, color]` 97 * Color is a `str` representing a color name or hexcode. 98 * See https://matplotlib.org/stable/users/explain/colors/colors.html 99 * ex. `0\t90\tblue` 100 """ 101 rescale_tri: bool = True 102 """ 103 Rescales track proportions so always a right isosceles triangle. 104 * https://byjus.com/maths/isosceles-right-triangle/ 105 """
Self-identity heatmap triangle plot options.
Colorscale for identity as TSV file.
- Format:
[start, end, color]- Color is a
strrepresenting a color name or hexcode. - See https://matplotlib.org/stable/users/explain/colors/colors.html
- Color is a
- ex.
0 90 blue
151@dataclass 152class LocalSelfIdentTrackSettings(LabelTrackSettings): 153 """ 154 Local self-identity plot options. 155 """ 156 157 colorscale: Colorscale | str | None = None 158 """ 159 Colorscale for identity as TSV file. 160 * Format: `[start, end, color]` 161 * Color is a `str` representing a color name or hexcode. 162 * See https://matplotlib.org/stable/users/explain/colors/colors.html 163 * ex. `0\t90\tblue` 164 """ 165 band_size: int = 5 166 """ 167 Number of windows to calculate average sequence identity over. 168 """ 169 ignore_band_size: int = 2 170 """ 171 Number of windows ignored along self-identity diagonal. 172 """
Local self-identity plot options.
Colorscale for identity as TSV file.
- Format:
[start, end, color]- Color is a
strrepresenting a color name or hexcode. - See https://matplotlib.org/stable/users/explain/colors/colors.html
- Color is a
- ex.
0 90 blue
269@dataclass 270class StrandTrackSettings(DefaultTrackSettings): 271 """ 272 Strand arrow plot options. 273 """ 274 275 DEF_COLOR = "black" 276 """ 277 Default color for arrows. 278 """ 279 scale: float = 50 280 """ 281 Scale arrow attributes by this factor as well as length. 282 """ 283 fwd_color: str | None = None 284 """ 285 Color of `+` arrows. 286 """ 287 rev_color: str | None = None 288 """ 289 Color of `-` arrows. 290 """ 291 use_item_rgb: bool = False 292 """ 293 Use `item_rgb` column if provided. Otherwise, use `fwd_color` and `rev_color`. 294 """
Strand arrow plot options.
337@dataclass 338class HORTrackSettings(DefaultTrackSettings): 339 """ 340 Higher order repeat plot options. 341 """ 342 343 sort_order: str = "descending" 344 """ 345 Plot HORs by `{mode}` in `{sort_order}` order. 346 347 Either: 348 * `ascending` 349 * `descending` 350 * Or a path to a single column file specifying the order of elements of `mode`. Only for split. 351 352 Mode: 353 * If `{mer}`, sort by `mer` number 354 * If `{hor}`, sort by `hor` frequency. 355 """ 356 mode: Literal["mer", "hor"] = "mer" 357 """ 358 Plot HORs with `mer` or `hor`. 359 """ 360 live_only: bool = True 361 """ 362 Only plot live HORs. Filters only for rows with `L` character in `name` column. 363 """ 364 mer_size: int = 171 365 """ 366 Monomer size to calculate number of monomers for mer_filter. 367 """ 368 mer_filter: int = 2 369 """ 370 Filter HORs that have less than `mer_filter` monomers. 371 """ 372 hor_filter: int = 5 373 """ 374 Filter HORs that occur less than `hor_filter` times. 375 """ 376 color_map_file: str | None = None 377 """ 378 Monomer color map TSV file. Two column headerless file that has `mode` to `color` mapping. 379 """ 380 use_item_rgb: bool = False 381 """ 382 Use `item_rgb` column for color. If omitted, use default mode color map or `color_map`. 383 """ 384 split_prop: bool = False 385 """ 386 If split, divide proportion evenly across each split track. 387 """ 388 split_top_n: int | None = None 389 """ 390 If split, show top n HORs for a given mode. 391 """ 392 393 split_fill_missing: str | None = None 394 """ 395 If split and defined sort order provided, fill in missing with this color. Otherwise, display random HOR variant. 396 * Useful to maintain order across multiple plots. 397 """ 398 399 split_sort_order_only: bool = False 400 """ 401 If split and defined sort order provided, only show HORs within defined list. 402 """ 403 404 bg_border: bool = False 405 """ 406 Add black border containing all added labels. 407 """ 408 409 bg_color: str | None = None 410 """ 411 Background color for track. 412 """
Higher order repeat plot options.
Plot HORs by {mode} in {sort_order} order.
Either:
ascendingdescending- Or a path to a single column file specifying the order of elements of
mode. Only for split.
Mode:
- If
{mer}, sort bymernumber - If
{hor}, sort byhorfrequency.
Monomer color map TSV file. Two column headerless file that has mode to color mapping.
Use item_rgb column for color. If omitted, use default mode color map or color_map.
If split and defined sort order provided, fill in missing with this color. Otherwise, display random HOR variant.
- Useful to maintain order across multiple plots.
297@dataclass 298class HOROrtTrackSettings(StrandTrackSettings): 299 """ 300 Higher order repeat orientation arrow plot options. 301 """ 302 303 live_only: bool = True 304 """ 305 Only plot live HORs. 306 """ 307 mer_filter: int = 2 308 """ 309 Filter HORs that have at least 2 monomers. 310 """ 311 arr_opt_bp_merge_units: int | None = 256 312 """ 313 Merge HOR units into HOR blocks within this number of base pairs. 314 """ 315 arr_opt_bp_merge_blks: int | None = 8000 316 """ 317 Merge HOR blocks into HOR arrays within this number of bases pairs. 318 """ 319 arr_opt_min_blk_hor_units: int | None = 2 320 """ 321 Grouped stv rows must have at least `n` HOR units unbroken. 322 """ 323 arr_opt_min_arr_hor_units: int | None = 10 324 """ 325 hor_len_Require that a HOR array have at least `n` HOR units. 326 """ 327 arr_opt_min_arr_len: int | None = 30_000 328 """ 329 Require that a HOR array is this size in bp. 330 """ 331 arr_opt_min_arr_prop: float | None = 0.9 332 """ 333 Require that a HOR array has at least this proportion of HORs by length. 334 """
Higher order repeat orientation arrow plot options.
Merge HOR units into HOR blocks within this number of base pairs.
Merge HOR blocks into HOR arrays within this number of bases pairs.
175@dataclass 176class BarTrackSettings(DefaultTrackSettings): 177 """ 178 Bar plot options. 179 """ 180 181 DEF_COLOR = "black" 182 """ 183 Default color for bar plot. 184 """ 185 186 color: str | None = None 187 """ 188 Color of bars. If `None`, uses `item_rgb` column colors. 189 """ 190 191 alpha: float = 1.0 192 """ 193 Alpha of bars. 194 """ 195 196 ymin: int | Literal["min"] = 0 197 """ 198 Minimum y-value. 199 * Static value 200 * 'min' for minimum value in data. 201 """ 202 203 ymin_add: float = 0.0 204 """ 205 Add some percent of y-axis minimum to y-axis limit. 206 * ex. -0.05 subtracts 5% of min value so points aren't cutoff in plot. 207 """ 208 209 ymax: int | Literal["max"] | None = None 210 """ 211 Maximum y-value. 212 * Static value 213 * 'max' for maximum value in data. 214 """ 215 216 ymax_add: float = 0.0 217 """ 218 Add some percent of y-axis maximum to y-axis limit. 219 * ex. 0.05 adds 5% of max value so points aren't cutoff in plot. 220 """ 221 222 label: str | None = None 223 """ 224 Label to add to legend. 225 """ 226 227 add_end_yticks: bool = True 228 """ 229 Add y-ticks showing beginning and end of data range. 230 """
Bar plot options.
Add some percent of y-axis minimum to y-axis limit.
- ex. -0.05 subtracts 5% of min value so points aren't cutoff in plot.
Maximum y-value.
- Static value
- 'max' for maximum value in data.
233@dataclass 234class LineTrackSettings(BarTrackSettings): 235 """ 236 Line plot options. 237 """ 238 239 position: Literal["start", "midpoint"] = "start" 240 """ 241 Draw position at start or midpoint of interval. 242 """ 243 fill: bool = False 244 """ 245 Fill under line. 246 """ 247 linestyle: str = "solid" 248 """ 249 Line style. See https://matplotlib.org/stable/gallery/lines_bars_and_markers/linestyles.html. 250 """ 251 linewidth: int | None = None 252 """ 253 Line width. 254 """ 255 marker: str | None = None 256 """ 257 Marker shape. See https://matplotlib.org/stable/api/markers_api.html#module-matplotlib.markers, 258 """ 259 markersize: int | None = None 260 """ 261 Marker size. 262 """ 263 log_scale: bool = False 264 """ 265 Use log-scale for plot. 266 """
Line plot options.
Marker shape. See https://matplotlib.org/stable/api/markers_api.html#module-matplotlib.markers,
108@dataclass 109class LabelTrackSettings(DefaultTrackSettings): 110 """ 111 Label plot options. 112 """ 113 114 DEF_COLOR = "black" 115 """ 116 Default color for label. 117 """ 118 119 color: str | None = None 120 """ 121 Label color. Used if no color is provided in `item_rgb` column. 122 """ 123 124 use_item_rgb: bool = True 125 """ 126 Use `item_rgb` column if provided. Otherwise, generate a random color for each value in column `name`. 127 """ 128 129 alpha: float = 1.0 130 """ 131 Label alpha. 132 """ 133 134 shape: Literal["rect", "tri"] = "rect" 135 """ 136 Shape to draw. 137 * `"tri"` Always pointed down. 138 """ 139 140 edgecolor: str | None = None 141 """ 142 Edge color for each label. 143 """ 144 145 bg_border: bool = False 146 """ 147 Add black border containing all added labels. 148 """
Label plot options.
415@dataclass 416class LegendTrackSettings(DefaultTrackSettings): 417 index: int | list[int] | None = None 418 """ 419 Index of plot to get legend of. 420 """
